GIANT VIRUS
Pallvi Slathia1*, Deepti Narang2, Rinmuanpuii Ralte3
1Assistant Professor, Department of Veterinary Microbiology, KCVAS, Amritsar, 143001, Punjab, India
2Principal Scientist-cum-Head, Department of Veterinary Microbiology, College of Veterinary Science, Guru Angad Dev Veterinary and Animal Sciences University, Ludhiana-141004, Punjab, India
3Associate Professor, Department of Veterinary Microbiology, KCVAS, Amritsar, 143001, Punjab, India
*Corresponding author email: pallvi.slathia@yahoo.com
Abstract: The most common infectious agents on earth are viruses, which are present in practically every environment. Only a few numbers of genes necessary for virus replication and capsid synthesis are present in the majority of viruses. They have a genome that is over 1 Mbp in size is much larger than the other known viruses and approximately 1,000 proteins. After making this initial discovery, there have been numerous reports of numerous viral families’ worth of giant virus strains. The complexity in giant viral proteomes includes features which are usually reserved for cellular organisms. Components of the translational machinery, DNA maintenance, and metabolic enzymes are among these novel roles.
Keywords: Giant virus, Nucleocytoplasmic large DNA viruses (NCLDV), Mimiviridae, Marseilleviridae, Pandoraviridae
Giant Viruses and its discovery:
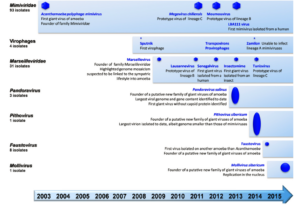
Giant viruses are also known as Girus, are double standard DNA viruses. The name giant virus is given to these viruses because of their extremely large genome size compared to other traditional viruses. These viruses encode hundreds of genes encoding proteins and can be visualized under light microscope unlike other viruses. They are classified under Nucleocytoplasmic Large DNA Viruses (NCLDV) (Brandes and Linial, 2019). Acanthamoeba polyphaga mimivirus (APMV), the first gigantic virus, was identified in 2003 (La Scola et al., 2003). Its size, which was comparable to microscopic bacteria or archaea, was unprecedented. APMV was visible under a light microscope, unlike any other virus that had previously been discovered (Xiao et al., 2005). Only ten years after its isolation was it identified as a virus after first being thought to be a bacterium (Abergel et al., 2015). Numerous other large viral species have been discovered following the initial discovery of APMV (Aherfi et al., 2016) (Figure 1). Although there are now many more representatives of giant viruses, the proportion of uncharacterized proteins in their proteomes is still remarkably high. Many of these uncharacterized proteins were also categorised as orphan genes (ORFans), meaning that no substantial sequence match was found.
Figure 1: Discovery of major representatives of giant viruses
Giant virus vs traditional viruses:
Size: Since a long time ago, viruses have only been strictly understood as microscopic infectious particles that can pass through 0.2 mm-pore filters and cannot be seen under a light microscope. In contrast, Megavirales virions (giant virus) range in size from 0.2 to 1.5 mm. As a result, for a very long time, Mimivirus and Pandoravirus virions were regarded as Gram-positive bacteria and parasitic endosymbionts, respectively. They also have giant genome structure.
Complexity: In terms of their nucleic acid and protein composition, giant viruses are more complex than “conventional” viruses. As a result, unlike the majority of other viruses, Megaviruses include both DNA and RNA, including messenger RNAs and transfer RNAs. Additionally, proteomics found dozens or perhaps hundreds of proteins inside giant virions, many of which are hypothetical proteins and some of which are involved in transcription and translation. These messenger RNAs and proteins could speed up the first stages of the replicative cycle, reducing the dependence of giant viruses on their hosts for replication compared to other viruses (Legendre et al., 2015).
Translation components: A peptide chain release factor, a GTP-binding elongation factor, translation initiation factors, and four aminoacyl-tRNA synthetases, some of which were proved to be functional and expressed, were discovered as a result of the discovery of Mimivirus (Raoult et al., 2004). Only one gene for a translation elongation factor had previously been discovered in Phycodnaviruses. Six transfer RNAs were also discovered. Then, genes for translational proteins and tRNA were found. This is a highly characteristic trait of these viruses (Claverie and Abergel, 2010).
Mobilome: Genetic components that can travel both within and between genomes are referred to as the mobilome. While they are uncommon in traditional virues, group I and II introns were frequently found in the conserved genes of giant viruses (La Scola et al., 2003). Additionally, transpovirons, a new type of transposable elements, were found in Mimiviruses; they are related to virus-associated plasmids seen in bacteria and archaea and depend on these large viruses for reproduction and spread. DNA transposable elements were also found in the genome of some giant viruses eg: P. salinus (Desnues et al., 2015).
Broad host spectrum: Giant virus such as Megaviruses infect a wide variety of cellular hosts, including invertebrates, mammals, amoebozoa, green algae and chromal-veolates, which are phylogenetically very distant from them. This is in contrast to viruses from other orders or families (Koonin and Yutin, 2010). Various protists, insects, and mammals, including humans, have been found to harbour Mimiviruses and Marseilleviruses. It has also been demonstrated that Mimivirus enters macrophages through a mechanism similar to phagocytosis, operating like bacteria. Giant viruses that infect amoebas also enter their host through phagocytosis (La Scola et al., 2003; Philippe et al., 2013). This differs with “conventional” virus entry processes, which require particular interactions with cell receptors (Ghigo et al., 2008).
Classification:
- Superfamily: Nucleocytoplasmic large DNA viruses (NCLDV).
- Family: Mimiviridae
- Family: Marseilleviridae
- Family: Pandoraviridae
- Genus: Pithovirus, Mollivirusand Faustovirus
Family- Mimiviridae: Acanthamoeba polyphaga mimivirus (APMV) is the first representative of this family with a capsid size of ~400nm capsid size. They have fibrils that are ~120–140 nm long 1.4 nm thick and genome length of 1.2 Mb containing 979 genes encoding proteins. They have aminoacyl tRNA synthetases and their genome encodes four different tRNAs and >75% ORFan genes. They were identified as viral particles by electron microscopy, then named Mimivirus for “Mimicking microbes (Sharma et al., 2016).
Family- Marseilleviridae: In 2007, Marseillevirus was isolated in Marseille, France. Acanthamoeba polyphaga marseillevirus (APMaV), its first member, was discovered in 2007 after being cultured on amoebae from water taken from a cooling tower in Paris, France. APMaV has a diameter of 250 nm and an icosahedral symmetry. Marseillevirus have dsDNA (350–380 kbp large) encoding 457 genes (Colson et al., 2013).
Family- Pandoraviridae: The virions are irregular ovoid. It has the largest viral genome size i.e., 2500 kbp with 2556 predicted proteins by its genome. More than 85% of its genome has no detectable sequence similarity to any other sequence in the public databases (Legendre et al., 2018).
Genus- Pithovirus: Pithovirus has circular, dsDNA and are also called as Zombie virus with 1500 nm by 500 nm in size and 610 kbp genome. The name Pithovirus, refers to big storage containers of ancient Greece known as pithoi. From a from a >30,000-year-old dated Siberian permafrost sample, Pithovirus sibericum was isolated (Legendre et al., 2015). It encodes 467 ORFs and translating to 467 proteins. It is the largest virus in terms of size found yet. Internally, its structure resembles a honeycomb. Recent isolation of Pithovirus massiliensis from sewage.
Genus- Mollivirus: The virion has an original spherical shape, a diameter of 500–600 nm. The ds DNA genome of M. sibericum is linear, 651 kb in length. Host encodes 523 proteins & histone homologs and HMG-like chromatin-associated proteins and has 65% are ORFans (Legendre et al., 2015).
Genus- Faustovirus: Faustovirus virions have an icosahedral capsid with a diameter of 200 nm. It was the first giant virus isolated on another free-living amoeba than Acanthamoeba. Its genome is a 461 kb long circular dsDNA with 451 proteins predicted. It also has genes encoding a ribosomal protein acetyl transferase, a bacteriophage tail fiber protein and two polyproteins shared with Asfarvirus (Reteno et al., 2015).
References:
Abergel, C., Legendre, M. and Claverie, J.M. (2015). The rapidly expanding universe of giant viruses: Mimivirus, Pandoravirus, Pithovirus and Mollivirus. FEMS Microbiological Reviews 39: 779–796.
Aherfi, S., Colson, P., LaScola, B. and Raoult, D. (2016). Giant Viruses of Amoebas: An Update. Frontiers in Microbiology 7: 349.
Brandes, N. and Linial, M. (2019). Giant Viruses—Big Surprises. Viruses 11: 404.
Claverie, J.M. and Abergel, C. (2010). Mimivirus: the emerging paradox of quasi-autonomous viruses. Trends Genetics 26: 431-437.
Colson, P., Pagnier, I., Yoosuf, N., Fournous, G. and La Scola, B. (2013). ‘‘Marseilleviridae’’, a new family of giant viruses infecting Amoebae. Archives in Virology 158: 915-20.
Desnues, C., La Scola, B., Yutin, N., Fournous, G., Robert, C., Azza, S., Jardot, P., Monteil, S., Campocasso, A., Koonin, E.V. and Raoult, D. (2012). Provirophages and transpovirons as the diverse mobilome of giant viruses. Proceedings of the National Academy of Sciences of the U S A 109: 18078-83.
Ghigo, E., Kartenbeck, J., Lien, P., Pelkmans, L., Capo, C., Mege, J.L. and Raoult, D. (2008) Ameobal pathogen mimivirus infects macrophages through phagocytosis. PLoS Pathogens 4: e1000087.
Koonin, E.V. and Yutin, N. (2010). Origin and evolution of eukaryotic large nucleo-cytoplasmic DNA viruses. Intervirology 53: 284-292.
La Scola, B., Audic, S., Robert, C., Jungang, L., de Lamballerie, X., Drancourt, M., Birtles, R., Claverie, J.M. and Raoult, D. (2003). A giant virus in amoebae. Science 299: 2033.
Legendre, M., Lartigue, A., Bertaux, L., Jeudy, S., Bartoli, J., Lescot, M., Alempic, J.M., Ramus, C., Bruley, C., Labadie, K., Shmakova, L., Rivkina, E., Coute, Y., Abergel, C. and Claverie, J.M. (2015). In-depth study of Mollivirus sibericum, a new 30,000-yold giant virus infecting Acanthamoeba. Proceedings of the National Academy of Sciences of the U S A 112: E5327-E5335.
Legendre, M., Fabre, E., Poirot, O., Jeudy, S., Lartigue, A., Alempic, J.M., Beucher, L., Philippe, N., Bertaux, L., Christo-Foroux, E., Labadie, K., Coute, Y., Abergel, C . and Claverie, J.M. (2018). Diversity and evolution of the emerging Pandoraviridae family. Nature 9: 2285.
Philippe, N., Legendre, M., Doutre, G., Coute, Y., Poirot, O., Lescot, M., Arslan, D., Seltzer V., Bertaux, L., Bruley, C., Garin, J., Claverie, J.M. and Abergel, C. (2013). Pandoraviruses: amoeba viruses with genomes up to 2.5 Mb reaching that of parasitic eukaryotes. Science, 341: 281-286.
Raoult, D., Audic, S., Robert, C., Abergel, C., Renesto, P., Ogata, H., La Scola, B., Suzan, M. and Claverie, J.M. (2004). The 1.2-megabase genome sequence of Mimivirus. Science, 306: 1344-50.
Reteno, D.G., Benamar, S., Khalil, J.B., Andreani, J., Armstrong, N., Klose, T., Rossmann, M., Colson, P., Raoult, D. and La Scola, B. (2015). Faustovirus, an Asfarvirus-Related New Lineage of Giant Viruses Infecting Amoebae. Journal of Virology 89: 6585–94.
Sharma, V., Colson, P., Pontarotti, Pierre. and Raoult, D. (2016). Mimivirus inaugurated in the 21st century the beginning of a reclassification of viruses. Current Opinion in Microbiology 31: 16–24.
Xiao, C., Chipman, P.R., Battisti, A.J., Bowman, V.D., Renesto, P., Raoult, D. and Rossmann, M.G. (2005). Cryo-electron microscopy of the giant Mimivirus. Journal of Molecular Biology 353: 493–496.