By-a)Mitek Taranga, B. L. Sainia,
b) Snehasmita Pandaa, Sachin,Tripura
a)Department of Animal Genetics and Breeding ,ICAR-Indian Veterinary Research Institute, Izzatnagar
b)Animal Nutrition Division, ICAR-National Dairy Research Institute, Karnal
Introduction:
A new technology called genomic selection is revolutionizing livestock breeding. Genomic selection is a type of marker-assisted selection in which genetic markers covering the whole genome are used so that all quantitative trait loci are in linkage disequilibrium with atleast one marker. Genomic selection is based on estimation of detailed associations between a very dense set of genetic markers (SNP-single nucleotide polymorphism) and phenotypes on a select group of animals: the reference population. The resulting prediction equations are then applied to SNP genotypes of the rest of the population to estimate their genomic breeding value (gEBV), without the need of additional phenotypes. The term genomic selection was first used by Meuwissen and coworkers in 2001.
The genomic selection revolution began with 2 developments. The first was the recent sequencing of the bovine genome, which led to the discovery of many thousands of DNA markers, in the form of SNP. The second development was the demonstration that it was possible to make very accurate selection decisions when breeding values were predicted from dense marker data alone.
Principle of genomic selection
The basic principle of genomic selection is the use of marker information for estimation of breeding value without having the information of gene location. There are two groups of animals: reference population with detailed phenotypes, and a large group without those phenotypes, also called the main population (candidates for selection). For genotypic information, all animals in reference population are genotyped for SNP markers of entire genome. In the reference population the associations between markers and phenotype are estimated and are used to obtain predictive equation. Then, those prediction equations are combined with the genotypes of the animals in the population (candidates for selection) to estimates their breeding values (gEBV), with only marker genotype information.
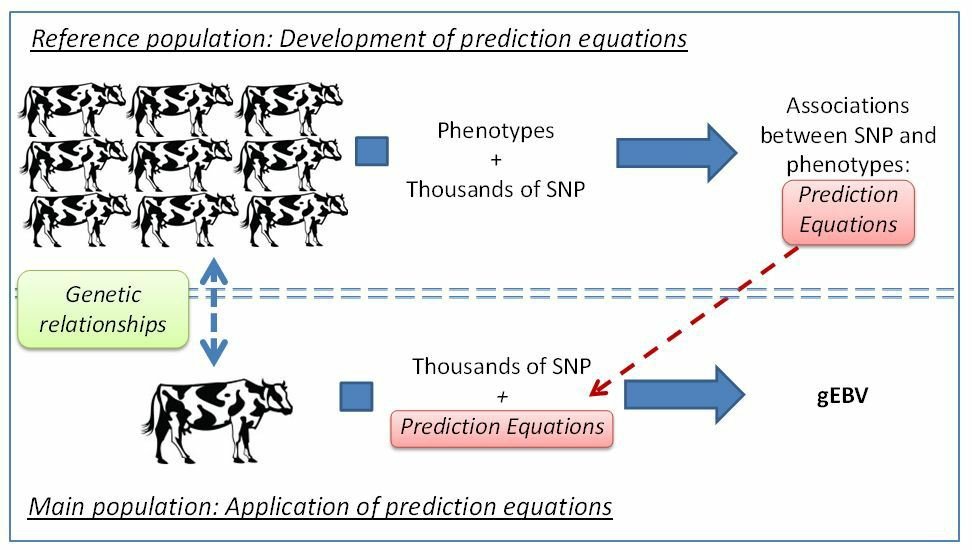
Fig 1: Principles of genomic selection. Top: a prediction equation is obtained from a reference population with phenotypes and genotypes; bottom: this prediction equation is used on candidates with genotypic information only.
Benefits of implementing genomic selection:
It increases the productivity of livestock much faster than traditional breeding methods, through increasing rate of genetic gain (yearly could be doubled) by decreasing generation interval.
It Makes progeny testing no longer necessary and slower the rate of inbreeding
It accuracy rate is more as compare to traditional approach
It Can be applied for the improvement of following traits:
1. That are very difficult or expensive to measure
2. For sex limited traits eg. milk production traits, egg production traits
3. For low heritability traits such as fertility or disease resistance like mastitis
4. To estimate the breeding value of very young animals, before they can produce a phenotype
Progress of genomic selection in livestock improvement:
In dairy cattle, SNP genotyping has been used to discover markers that will improve the reliability of traits associated with milk production, cow health, and cow conformation.
A number of SNP chips have been developed for bovine which helps in genomic selection. Few examples are: Bovine 50K (Illumina), 75K (Geneseek), 700K (affymetrix).
INDUSCHIP from indigenous cattle sample has been developed by National Dairy Development Board (NDDB).
The reliability and accuracy of genomic prediction in dairy cattle exceeds 0.8 for production traits and 0.7 for fertility and other traits.
When using genomic selection generation intervals were suggested to drop from around 5 to 6 years in traditional dairy cattle breeding programs to around 1.5 years.
NDDB has now developed world’s first complete parent-wise genome assembly of buffalo named as ‘NDDB ABRO Murrah’. This genome assembly of riverine buffaloes would provide more insights about buffalo genome and will helps to contribute more in genomic selection.
In pig, genomic selection could possibly reduce generation interval up to 25% and improve litter size much faster as compared to traditional methods.
Genomic selection also successfully increased the genetic gain of the trait: post weaning mortality in pig by increasing the accuracy of gEBV up to 50%. It has been made possible by selection with high accuracy to increase the number of teats to ensure that sows can nurture more number of live piglets.
Soon after the release of the draft chicken genome sequence in 2004, the EW Group started to develop its first SNP panel. Due to rapid technological advances, the chip density increased from 6Kto 12K, to 42Kand recently to 60K SNPs.
Genomic selection was implemented by Hy-Line International in some lines of layers using the newly developed 60K SNP chip and the first genomic selected birds was generated, descendants enter the market as commercial birds in 2015.
Constraints in implementation:
Require large size of reference population with thousands of animals measured for phenotype and genotype.
Increase cost as larger population phenotyping and detailed genotyping obviously is more expensive.
How the farmers can play a role in implementation genomic selection?
Since the genomic selection method to be effective, is very demanding in terms of both number of individuals genotyped and association of genomic regions with a range of phenotypes or performance levels. Therefore maintaining regular recording of reliable and accurate data of high economic (milk, SNF etc) traits and their sharing to national genetic evaluation system (NGES) is very much needed that can be easily possible through farmers participation. The NGES can records those data in real time; so that large reference population of good phenotyping producing animals can be easily made. That’s why farmer’s association and dairy organization can play a great role in this way for early and accurate implementation of genomic selection.
Conclusions:
Genomic selection is a potential tool to increase animal production through improving genetic gain. Strong evolutions have started, including reduction in genotyping costs, phenotyping strategies for new traits, approaches for the creation or the replacement of reference population etc. With these research progresses, and farmers participation in data recording genomic selection will become an efficient tool for the production of elite animals in all livestock species.


